As a webinar relating to natural science and information systems, I picked the Webinar from the ScieneMagazine (2018): http://www.sciencemag.org/custom-publishing/webinars/addressing-reproducibility-crisis-validating-antibodies-life-science , which is a video recording of the live event from 11th April 2018. The topic is interesting because data quality and integrity are highly important as organisations need to rely on it.
Summary
After we are invited to share the webinar via social media, the two professors M. Uhlen and C. Williams go over the three agenda points:
Upfront, the “Human Protein Atlas” https://www.proteinatlas.org is presented, which allows retrieval and submission of data about human proteins with antibodies from everybody. The information that can be received contains up-to-day data for the antibody discovery, mostly supported by microscopic pictures of the discoveries/objects. The first topic for antibody validation highlights the need to establish ground rules to have accurate data, reducing unwanted errors.
The second subject shows the validation case study around ER B antibody receptors and how this became problematic as reproduction of the results failed in 12 out of 13 cases, see figure 1. This led to sunken cost and an inadequate basis for clinical trials, estimating 28,2 US Dollar to be spent on irreproducible antibody research, the Merck AG from Darmstadt, Germany even called it a horror show.
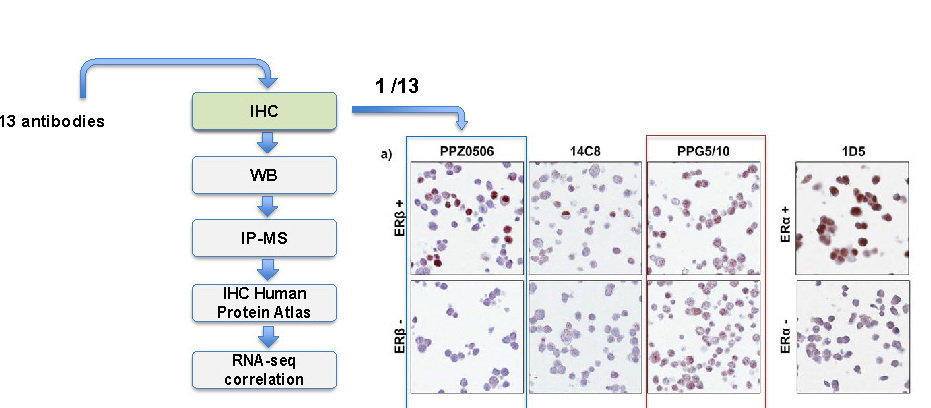
Figure 1: 12 out of 13 ERβ antibodies were inadequate on controls
Source: Adapted from: Andersson et al., Nature Communications (2017)
The third section deals with an enhanced validation proposal consisting of five “pillars”. Recent updates on the “Protein Atlas” have proofed the existing antibodies with this enhance validation method, thus giving the data in the system more accurate. These newly validated ensure security that the data is not corrupted, despite the open access/submission system it is based on.
Critic / Opinion
Protein Atlas: This makes data sharing and consumption thought the field possible.
On Topic 1 – Errors: Unwanted noise should be reduced (Quince, Lanzen, Davenport, & Turnbaugh, 2011) to establish a trustworthy data base, in this case to avoid the three standard issues in reliable antibody use (Target binding, cross-reactivity and reproducibility).
On Topic 2 Practice: The conclusion is that validation of research results is critical but time-consuming; However, the webinar does not specify a “best practice” on the (minimal) ratio of time on research and time to be spent on validation.
On Topic 3 – Enhance checks: The Article highlights how data integrity should be checked in their field, which was presented clearly. In IS/information technology (IT), when minor variations/unclarities cause issues to the organisation/business relying on it, they need to protect themselves from such issues as well:
Both side (IT/IS) need to have appropriate strategies, algorithm and hardware in place to avoid data glitches, which is not highlighted and somewhat taken for granted in the article. For example, servers for data store and computation should have error protection mechanisms, starting with memory error checking, even if virtualized (Yoon & Erez, 2011).
The presentation (and moderation) was done very well, the topics integrated seamlessly and were well illustrated for better digestion.
The webinar and the “Protein Atlas” shows that information systems are at the heart of science, which tends to advance with open access and therefore public review.
It must be noted that noise, as mentioned here by the author, is a negative, noise in IS can also be used to an advantage like shown in the paper Noise-Enhanced Information Systems by (Chen, Varshney, & Varshney, 2014).
References
Chen, H., Varshney, L. R., & Varshney, P. K. (2014). Noise-Enhanced Information Systems. Proceedings of the IEEE ( Volume: 102, Issue: 10, Oct. 2014 ) (pp. 1607 – 1621). IEEE. doi:https://doi-org.salford.idm.oclc.org/10.1109/JPROC.2014.2341554
Quince, C., Lanzen, A., Davenport, R. J., & Turnbaugh, P. J. (2011). Removing Noise From Pyrosequenced Amplicons. BMC Bioinformatics, 1471. doi:https://doi.org/10.1186/1471-2105-12-38
Science Magazine. (2018, June 12). WEBINAR TECHNOLOGY Addressing the reproducibility crisis: Validating antibodies for life science research. Retrieved from Science Mag: http://www.sciencemag.org/custom-publishing/webinars/addressing-reproducibility-crisis-validating-antibodies-life-science
Yoon, D., & Erez, M. (2011). VIRTUALIZED ECC: FLEXIBLE RELIABILITY IN MAIN MEMORY. Ieee Micro, 31(1),, pp. 11-19.
